
EMSA竞争实验
本英文及附属网页是我们美国网站的原稿内容。因为我们的中文文案常被盗用,就不准备翻译成中文了。如有任何问题,请与我们联系,谢谢理解。
After the shift bands of
protein-nuclear acid complexes are observed in EMSA, Two
common approaches, Competitive EMSA or/and Supershift EMSA
(detail in other section), are used to determine the
specificity of protein and nuclear acid interaction.
1. What is the Competitive
EMSA? Why need it?
Competitive EMSA is the most
common test of specificity. Prior to the addition of p32-,
biotin- or fluorescent dye-probes, a 50-100 fold molar
excess of unlabeled competitor DNA is added to the reaction
mix. Individual reactions are performed with
oligonucleotides containing the target DNA sequence and
oligonucleotides which have been specifically mutated within
the target sequence. Specific binding is indicated by a loss
of factor binding to the biotin- or fluorescent dye-probes.
The alteration of conserved bases within the binding site
can abolish the ability of a transcription factor to bind to
its cognate DNA. Thus for site-directed mutant competitions,
binding of the labeled probe is preserved. In
addition, competitions should also be performed with well
characterized consensus DNA sequences specific for the
factor of interest.
2. How to design specific competitive or mutant
probes?
Usually, a specific
competitive probe is unlabeled (referred as cold probe) and
has the same sequence (or at least with the same binding
domain) as that of labeled probes. The mutant probe is also
unlabeled, and has the sequence as that of labeled probes,
but the binding domain has been mutant with one or more
bases.
Designing a mutant probe
follows these rules: 1) muting one or more bases in the
binding domain of consensus sequences. 2) Usually, changing
AT to GC, or vice versa. 3) checking mutant probes to
avoid forming another binding domain by making
adjustment. 4) Verifying mutant-probes with EMSA.
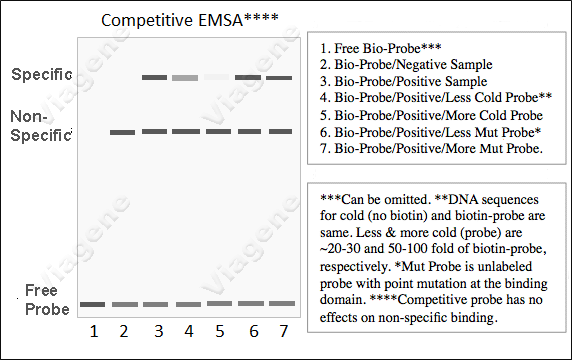
3. How to perform Competitive
EMSA?
In order to get an expecting result from Competitive EMSA, the experiments should consider following points:
(1) The cell or tissue
extracts should have activated proteins/factors enabled to
bind to DNA or RNA probes.
(2) Shifted bands of
protein-nuclear acid complexes must be detected in
conventional EMSA.
(3) Competitive or mutant probes should
be added to reaction mixture before the reaction of proteins
and DNA/RNA.
(4) Nuclear proteins used for EMSA are
isolated with a high-salt buffer. High volume of nuclear
extracts may interrupt protein/nucleic acid interaction.
(6) The radio of competitive probes/labeled probes varies
significantly among different DNA/RNA binding proteins. The
ratio of 1:20 may cause serious competition for one protein
factor, but as high up to ratio of 1:200 may require to
producing visible competition for another protein factor.
(7) A classic Competitive EMSA may consider to include
these reactions: 1) A sample 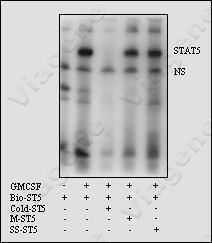 without activated target
proteins (negative sample) + labeled probes, 2) A sample
with activated target proteins + labeled probe (positive
sample), 3) positive sample + labeled probe + low dose of
specific competitors (competitive low), 4) positive sample +
labeled probe + high dose of specific competitors
(competitive high), 5) positive sample + labeled probe + low
dose of mutant probes (mutant low), 6) positive sample +
labeled probe + low dose of specific competitors (mutant
high).
without activated target
proteins (negative sample) + labeled probes, 2) A sample
with activated target proteins + labeled probe (positive
sample), 3) positive sample + labeled probe + low dose of
specific competitors (competitive low), 4) positive sample +
labeled probe + high dose of specific competitors
(competitive high), 5) positive sample + labeled probe + low
dose of mutant probes (mutant low), 6) positive sample +
labeled probe + low dose of specific competitors (mutant
high).
4. Interpretation
of Competitive EMSA
5. Common problems with Competitive EMSA
(1) No competition with unlabeled probes, which could be caused by
1) the complexes are formed by
non-specific interaction. 2) no enough competitive probes are used.
The radio of competitors/labeled probes varies among
different binding proteins; the ratio of 1:20 may
cause serious competition for one protein factor, but as
high up to the ratio of 1:200 may require to producing visible
competition for other protein factors. 3) amount of labeled
probes is used too much.
(2) Both target and non-specific
bands disappear when unlabeled probes are used, which could be
caused by too much competitive probes being used.
(3) Mutant probes cause significant
competition, which could be caused by 1) the point-mutation of
binding domain is not good enough to prevent the binding
of the probe to protein factors. 2) the amount of
mutant probes is used too much.
In any case, if
non-specific bands become weak or disappearing, indicating
amounts of competitive or mutant probes are used too much.
Related Products
| Products/Cat# | Description | Pack | Prices |
|---|---|---|---|
| See related products | General non-radioactive EMSA kits without probes & membrane | vary | vary |
| See related products | Complete non-radioactive EMSA kits with probe & membrane | vary | vary |
| See related products | Non-radioactive, ready probes. | vary | vary |
| See related products | Custom non-radioactive EMSA probe | vary | vary |
| See related products | EMSA positive/negative controls | vary | vary |
| See related products | EMSA related chemicals and reagents | vary | vary |

 客服一号
客服一号
